
Most of these techniques are specifically designed for a certain enzyme or enzyme class, which limits their applicability. Such approaches include the use of genetic and pharmacologic perturbations 11, substrate-trapping mutants 12, affinity purification-mass spectrometry 13, utilizing peptide 14 or protein arrays 15, tagging the client proteins by substrate analogs using engineered enzymes 16 and peptide immunoprecipitation 17 or the employment of sophisticated computational tools 18.
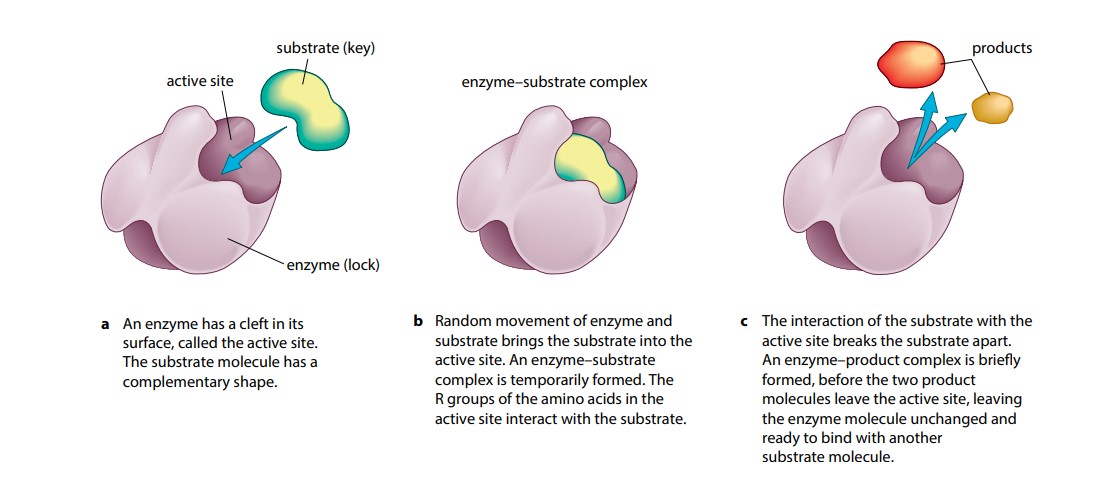
Most existing techniques used for identifying specific substrates are enzyme-specific, labor-intensive, and often not straightforward. Sometimes, however, there are too many known substrates, and an intelligent prioritization approach is needed for further exploration. The deficit of information on the physiological substrates of enzymes hampers the development of effective therapeutics, e.g. Characterization of enzyme–substrate associations is essential for our understanding of cell biology and disease mechanisms moreover, many high-throughput drug screening assays rely upon modified substrates as a readout 7, 8. However, there is still a lack of the methods for identifying substrates undergoing structural changes due to modifications 4, 5, 6.

Not surprisingly, mechanisms and kinetics of protein modifications have become a vibrant research area. Transient modulation of protein post-translational modifications (PTMs) controls numerous cellular processes by inducing a host of downstream effects, such as changes in protein function, stability, interactions, hemostasis, localization, and cellular diversification 3. Only in human genome, an estimated 1089 non-metabolic enzymes are present 2, including for example more than 500 putative kinases.

Many of these enzymes catalyze the modifications of protein substrates. One of the most comprehensive enzyme databases BRENDA comprises >9 million protein sequences and encompasses 6953 classes of enzyme-catalyzed reactions 1.
Nature Communications volume 12, Article number: 1296 ( 2021)Īt least a third of all proteins possess enzymatic activity. System-wide identification and prioritization of enzyme substrates by thermal analysis


 0 kommentar(er)
0 kommentar(er)
